
Plasma Epstein-Barr virus DNA analysis for personalised management of nasopharyngeal carcinoma—current opportunities and challenges
Introduction
In endemic nasopharyngeal carcinoma (NPC), the close association with Epstein-Barr virus (EBV) (1,2) has led to the development of the various virus-related biomarkers. Among which, plasma EBV DNA is the most established biomarker of NPC (3). Plasma EBV DNA is well proven to be highly sensitive and specific for NPC and has demonstrated its clinical utility in all stages of cancer management, from screening to prognostication and monitoring of recurrence. In this review, we would summarise the clinical utility of plasma EBV DNA analysis for NPC management and discuss its potential to guide personalised treatment. In addition, we would review our recent knowledge of the molecular characteristics of plasma EBV DNA from NPC samples and the diagnostic implications.
Plasma EBV DNA as a tumour biomarker
The development of the real-time quantitative polymerase-chain-reaction (qPCR) assay for detection of EBV DNA was first described by our group in 1999 (3). Using the qPCR assay, we have shown that EBV DNA was present at high concentrations in the plasma of NPC patients. Importantly, as a tumour biomarker, plasma EBV DNA exhibited a positive linear correlation with tumour burden. In human patients, plasma EBV DNA concentration correlates with both clinical tumour stage (3-5) and anatomical tumour volume (including primary tumour and regional lymph nodes) measured by MRI volumetric analysis (6). In the mouse model, again, a positive relationship was observed between the NPC tumour xenograft mass and plasma EBV DNA concentrations (7).
The concentration of EBV DNA in the circulation is determined by its release from NPC cancer cells and the in vivo clearance. The release of EBV DNA is in turn determined by the cancer cell population and its turnover. Regarding the in vivo clearance, we have previously studied the clearance kinetics of plasma EBV DNA in the surgical treatment model (i.e., patients with recurrent NPC receiving nasopharyngectomy) (8). It was shown that the clearance followed a first-order decay kinetics with a short median half-life of about 2 hours. Given the rapid clearance of plasma EBV DNA (or circulating DNA in general), its quantitative level indeed reflects the tumour burden in an almost real-time manner. Therefore, plasma EBV DNA measured at the different time points with reference to the treatment regime have different biological implications. All the pre-, mid- and post-treatment levels of EBV DNA were shown to carry prognostic values for NPC. In a recent systematic review, Lee et al. (9) has summarized the timing of measurements of plasma EBV DNA as an NPC tumour biomarker in the different studies reported.
Clinical utility
For prognostication in patients with an established diagnosis of NPC
Pre-treatment plasma EBV DNA
Pre-treatment level could provide a molecular indication of the tumour load, in addition to the conventional anatomy-based tumour-node-metastasis (TNM) staging system. Importantly, pretreatment level was also shown to be a prognostic factor independent of cancer stage for survival on multivariate analysis (10-12) and a predictive factor of local recurrence and distant metastasis (10). Remarkably, this independent prognostic value exists for both early- and advanced-stage NPC. For advanced-stage NPC patients (11), worse prognosis was reported in those with higher pretreatment levels and they had more inferior overall and relapse-free survival. Similarly, for early-stage (stage I and II) NPC patients, those with higher EBV DNA levels had a poorer survival similar to that of stage III disease, and those with lower levels had a better survival similar to that of stage I disease (12). With all the evidence, it has been proposed to incorporate this molecular biomarker into the current anatomy-based TNM staging system. To illustrate, two recent studies (13,14) have demonstrated the additional value of better risk stratification power by combining plasma EBV DNA and TNM staging analysis in a recursive-partitioning analysis (RPA) model.
Post-treatment plasma EBV DNA
After completion of treatment of curative intent, plasma EBV DNA is expected to drop below a detectable level. Any detectable levels of plasma EBV DNA detected after treatment may imply failure of complete tumour eradication and residual disease. We have previously analysed 170 NPC patients in a prospective study and have shown that a high post-treatment EBV DNA level was predictive of a higher risk of recurrence and associated with a poorer prognosis (both progression-free and overall survival) (15). Furthermore, we have recently demonstrated improved risk stratification and better survival prediction by integrating the post-treatment EBV DNA level and TNM stage using the recursive-partitioning analysis in multiple sample cohorts (16).
Mid-treatment plasma EBV DNA
During the standard fractionated radiotherapy treatment course, mid-treatment level could reflect the tumour burden at the corresponding time-point and could therefore be used to imply the interim response to treatment and tumour radiosensitivity. We have previously studied the kinetics of plasma EBV DNA in NPC patients during radiotherapy through serial blood sampling (17). An initial rise in plasma EBV DNA was noted as a result of treatment-related cancer cell death, which was then followed by a decline in the level to reflect the tumour shrinkage. Subsequently, we have evaluated the prognostic value of a single measurement of mid-treatment plasma EBV DNA (at 4 weeks of chemoradiotherapy/radiotherapy) in a prospective study (18). Patients with detectable mid-treatment plasma EBV DNA had a poorer prognosis (higher risk of distant failure and worse progression-free and overall survival). It is important to note that, on multivariate analysis, mid-treatment EBV DNA was the only significant prognostic factor while neither pre-treatment EBV DNA nor tumour stage was significant.
To further extend the concept, Lv et al. (19) analysed plasma EBV DNA at multiple time-points during treatment in a group of patients with locally advanced NPC receiving induction chemotherapy (in addition to the concurrent chemoradiotherapy regime). Based on the serial change in the biomarker (i.e., molecular response), they devised a model to classify patients into 4 subgroups, namely, early, intermediate and late responders and treatment-resistant groups. Better risk prediction and prognostication were demonstrated using the proposed classification system compared to the plasma EBV DNA measurement at any single time-point.
For surveillance of recurrence
Plasma EBV DNA could be used as a blood-based surveillance tool for detection of recurrent NPC, in adjunct to endoscopy and magnetic resonance imaging (20-22). One benefit of a regular plasma EBV DNA testing for surveillance is that that the rise in the level could be detected prior to a symptomatic presentation by weeks to months (23). However, it is worth noting that plasma EBV DNA is more effective in picking up distant metastatic relapse than local recurrence. As reported in our case-control study (24), the sensitivity for detection of stage I–II tumour recurrence was 42% only and that for stage III–IV recurrence was 83%.
For screening among asymptomatic individuals
To prove that plasma EBV DNA is an effective screening biomarker of NPC, it is crucial to show that NPC could be readily identified at the pre-symptomatic stage through plasma EBV DNA testing. Therefore, in our prospective territory-wide study (25), we have recruited more than 20,000 asymptomatic Chinese middle-aged men who were then subject to PCR-based plasma EBV DNA testing. In this study, about 70% of screen-detected NPC were early-stage disease (stage I–II), in contrast to the only 20% among symptomatic cases according to the local cancer registry (26). The early cancer detection was also shown to be associated with a survival benefit. These screen-detected NPC patients enjoyed a more superior 3-year progression-free survival compared to symptomatic patients from a historical cohort. All these findings supported the utility of plasma EBV DNA for screening NPC.
Recent developments
To guide personalised NPC treatment
The treatment for NPC is an evolving paradigm (27). The current treatment backbone is radiotherapy, while concurrent chemoradiotherapy is considered in non-metastatic stage II–IV NPC (anatomic staging) with evidence supported by the Meta-Analysis of Chemotherapy in Nasopharynx Carcinoma (MAC-NPC) collaborative group (28). To further improve the survival outcome, different research groups have investigated treatment intensification, for example, through addition of induction or adjuvant chemotherapy (29). However, even patients with the same tumour stage are heterogenous and would have diverse clinical outcome. To illustrate, over 50% of patients with stage III or IV disease did not have recurrence even without adjuvant chemotherapy (30) and therefore treatment intensification in any form may seem unnecessary for these patients. At the same time, about 20% of patients with stage II disease would recur under the current treatment recommendation without adjuvant chemotherapy (30). Therefore, researchers are exploring the use of plasma EBV DNA for escalation (or de-escalation) of treatment on the basis that plasma EBV DNA is an independent prognosticator for disease recurrence and survival as discussed above. It was hoped that plasma EBV DNA could better stratify patients within the same tumour stage into the high-risk group for treatment intensification and low-risk group for sparing of additional treatment.
Chan et al. have recently reported the result of the first biomarker-driven randomized controlled trial (NPC0502) that was aimed to evaluate the use of post-treatment plasma EBV DNA analysis for risk stratification and guiding adjuvant chemotherapy (31). However, among patients with detectable post-treatment EBV DNA which was regarded as the high-risk group, there was no statistically significant difference in relapse-free survival between the treatment arm (use of adjuvant cisplatin and gemcitabine) versus the observation arm. There are several hypotheses for the negative finding proposed by the research group, including the choice of the same chemotherapeutic agent (therefore ineffective to an already resistant clone) and the compliance to adjuvant treatment.
There are other ongoing studies that continues to explore the use of plasma EBV DNA for treatment guidance. The NRG-HN001 study (NCT02135042) would investigate the utility of post-treatment plasma EBV DNA to guide adjuvant chemotherapy after addressing the issues identified in the NPC0502 trial mentioned above. In the EP-STAR study (NCT04072107), the research group would use the classification proposed by Lv et al. (19) based on mid-treatment EBV DNA clearance to guide the use of additional chemotherapy or immune checkpoint inhibitor.
To fully realize the potential of plasma EBV DNA analysis for treatment guidance, it is important to understand and evaluate the performance parameters (32) of the PCR assay being adopted, including limit of detection, limit of quantification, linearity of the assay across the measuring range, precision and reproducibility. These parameters have to be interpreted in the clinical context of how the biomarker is used. For example, when it is measured at a post-treatment time-point to infer the presence of subclinical residual disease, the limit of detection of the assay will affect the sensitivity for detection. In contrast, if a quantitative threshold is used for risk stratification, the limit of quantification, linearity of the assay across the measuring range, precision and reproducibility needs to be adequately evaluated.
As reported in an international collaborative project (33) involving us and other laboratories, there is a low interlaboratory concordance of EBV DNA results by different assays. There are a number of factors that could lead to variability in EBV DNA quantitation by different assays, including biological source, extraction and purification methods of EBV DNA (or plasma DNA in general), PCR reagents, technique and design (amplicon length, target gene and target sequence). In the collaborative study, we have specifically identified that the assay calibrator is one major factor that contributes to interlaboratory variation in the EBV DNA results. Assay harmonization is necessary to improve the interlaboratory concordance. Such work will allow direct comparison of quantitative levels from different assays and facilitate multi-centered trials for patient recruitment and result generalizability (34). In addition, assay harmonization across different laboratories would promote formulation of clinical practice guidelines (35) for wider clinical utility.
To enhance NPC screening performance
The conventional real-time qPCR assay yields quantitative readouts of EBV DNA (larger than the amplicon size) in a sample. Recently, we have revealed the molecular characteristics of plasma EBV DNA in NPC patients with diagnostic implications for the screening utility (Figure 1).
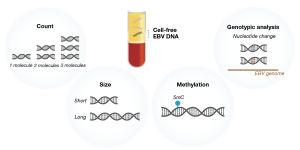
The benefits of NPC screening with PCR-based testing of plasma EBV DNA have been demonstrated in our prospective screening study (25). Within a screening population, about 5% of the people do not have NPC and yet they have detectable levels of plasma EBV DNA by PCR-based testing (36). We therefore adopted a two time-point testing protocol, that is, subjects were defined as screen-positive if they were positive for plasma EBV DNA both at recruitment and at re-test (4 weeks later). Such arrangement was based on the hypothesis that these non-NPC subjects harbour detectable levels of EBV DNA as a result of viral reactivation (37) and would have cleared the viral DNA on re-testing. This testing arrangement was shown to reduce the number of subjects with false positive results and therefore improve the specificity (25).
We have subsequently studied the molecular characteristics of plasma EBV DNA between NPC and non-NPC subjects by next-generation sequencing and discovered the differentiating quantitative and size profiles (38). NPC patients were found to have higher quantitative levels of plasma EBV DNA reads on sequencing. Regarding the size, plasma EBV DNA from NPC samples was shown to exhibit the characteristic nucleosome-associated size profile, with a modal peak at 166bp (that corresponds to mononucleosomal size) (39). In contrast, plasma EBV DNA from non-NPC samples did not have such nucleosomal size profile. Such difference was exploited to develop the size-based analysis of plasma EBV DNA for differentiating NPC and non-NPC subjects. Indeed, the size feature is one of the ‘fragmentomics’ markers of circulating DNA that we recently advocate (40). Fragmentomics refers to the study of non-random fragmentation process of circulating DNA and analysis of these fragmentomics markers (40-44) could yield important biological and diagnostic information.
In addition, we have recognized the differential methylation profiles of plasma EBV DNA between NPC and non-NPC subjects (45). Integrating the quantitative, size and methylation analysis of plasma EBV DNA was shown to substantially improve the diagnostic performance for NPC detection in screening. The combined analysis was shown to yield a modelled positive predictive value (i.e., 35.1%) which is three times of that by PCR-based two time-point testing protocol (i.e., 11.0%). Also, the combined molecular analysis of plasma EBV DNA allows a single time-point testing to achieve the high PPV. Such improvement in the diagnostic performance would reduce the number of screening participants for further confirmatory investigations.
Separately, we have recently reported the feasibility of EBV genotypic analysis through sequencing analysis of plasma DNA samples (46). A NPC risk score model for cancer prediction was developed based on the EBV genome-wide single nucleotide variant (SNV) profile. With the recent reports on the recognition of high-risk NPC-associated EBV variants (47,48), our NPC risk score analysis could be of potential use for stratifying screening subjects into different risk groups based on the viral variant profile. Different screening strategies may be adopted for the different risk groups. As an example, more frequent screening would be recommended for those with a high-risk score.
Conclusions
We have previously reviewed the biological properties of plasma EBV DNA and suggested that it could serve as an archetypal model to understand the biology of circulating tumour DNA (ctDNA) in general (49). The clinical applications of plasma EBV DNA analysis in NPC could indeed provide a good model to realise the full clinical potential of ctDNA analysis for other types of cancer also. Similar to plasma EBV DNA, ctDNA is now actively investigated for its clinical utility in screening, prognostication and surveillance of recurrence. We would envision that the plasma EBV DNA model could provide insights into solving the challenges associated with ctDNA analysis.
Acknowledgments
Funding: This work was supported by the Research Grants Council of the Hong Kong SAR Government under the Theme-based research scheme (T12-401/16-W), a collaborative research agreement from Grail, and the Innovation and Technology Fund under the InnoHK Initiative and the Vice Chancellor’s One-Off Discretionary Fund of The Chinese University of Hong Kong (VCF2014021). YMDL is supported by an endowed chair from the Li Ka Shing Foundation.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maria Li. Lung, Lawrence S. Young) for the series “NPC Biomarkers” published in Annals of Nasopharynx Cancer. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://anpc.amegroups.com/article/view/10.21037/anpc-21-11/coif). The series “NPC Biomarkers” was commissioned by the editorial office without any funding or sponsorship. WKJL holds equity in, and served as a consultant (from Feb 2018 to Jan 2019) to Grail. WKJL filed multiple patent applications on circulating nucleic acids analysis for cancer diagnostics. YMDL is a scientific co-founder, shareholder, scientific advisory board member and consultant of, and receives research support from Grail. YMDL is a founder, shareholder and board member of the Take2 Group of companies and DRA Limited, and an advisor of Decheng Capital. YMDL filed multiple patent applications on circulating nucleic acids analysis for cancer diagnostics. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 2016;16:789-802. [Crossref] [PubMed]
- Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006;15:1765-77. [Crossref] [PubMed]
- Lo YMD, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188-91. [PubMed]
- Lo YMD, Leung SF, Chan LY, et al. Plasma cell-free Epstein-Barr virus DNA quantitation in patients with nasopharyngeal carcinoma. Correlation with clinical staging. Ann N Y Acad Sci 2000;906:99-101. [Crossref] [PubMed]
- Fan H, Nicholls J, Chua D, et al. Laboratory markers of tumor burden in nasopharyngeal carcinoma: a comparison of viral load and serologic tests for Epstein-Barr virus. Int J Cancer 2004;112:1036-41. [Crossref] [PubMed]
- Ma BBY, King A, Lo YMD, et al. Relationship between pretreatment level of plasma Epstein-Barr virus DNA, tumor burden, and metabolic activity in advanced nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 2006;66:714-20. [Crossref] [PubMed]
- Chan KCA, Chan ATC, Leung SF, et al. Investigation into the origin and tumoral mass correlation of plasma Epstein-Barr virus DNA in nasopharyngeal carcinoma. Clin Chem 2005;51:2192-5. [Crossref] [PubMed]
- To EW, Chan KCA, Leung SF, et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 2003;9:3254-9. [PubMed]
- Lee AWM, Lee VHF, Ng WT, et al. A systematic review and recommendations on the use of plasma EBV DNA for nasopharyngeal carcinoma. Eur J Cancer 2021;153:109-22. [Crossref] [PubMed]
- Lo YMD, Chan ATC, Chan LY, et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res 2000;60:6878-81. [PubMed]
- Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461-70. [Crossref] [PubMed]
- Leung SF, Zee B, Ma BBY, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol 2006;24:5414-8. [Crossref] [PubMed]
- Guo R, Tang LL, Mao YP, et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma. Cancer 2019;125:79-89. [Crossref] [PubMed]
- Lee VH, Kwong DL, Leung TW, et al. The addition of pretreatment plasma Epstein-Barr virus DNA into the eighth edition of nasopharyngeal cancer TNM stage classification. Int J Cancer 2019;144:1713-22.
- Chan ATC, Lo YMD, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 2002;94:1614-9. [Crossref] [PubMed]
- Hui EP, Li WF, Ma BBY, et al. Integrating postradiotherapy plasma Epstein-Barr virus DNA and TNM stage for risk stratification of nasopharyngeal carcinoma to adjuvant therapy. Ann Oncol 2020;31:769-79. [Crossref] [PubMed]
- Lo YMD, Leung SF, Chan LY, et al. Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res 2000;60:2351-5. [PubMed]
- Leung SF, Chan KCA, Ma BBY, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol 2014;25:1204-8. [Crossref] [PubMed]
- Lv J, Chen Y, Zhou G, et al. Liquid biopsy tracking during sequential chemo-radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat Commun 2019;10:3941. [Crossref] [PubMed]
- Li WF, Zhang Y, Huang XB, et al. Prognostic value of plasma Epstein-Barr virus DNA level during posttreatment follow-up in the patients with nasopharyngeal carcinoma having undergone intensity-modulated radiotherapy. Chin J Cancer 2017;36:87. [Crossref] [PubMed]
- Hsu CL, Chan SC, Chang KP, et al. Clinical scenario of EBV DNA follow-up in patients of treated localized nasopharyngeal carcinoma. Oral Oncol 2013;49:620-5. [Crossref] [PubMed]
- Lee VH, Kwong DL, Leung TW, et al. Prognostication of serial post-intensity-modulated radiation therapy undetectable plasma EBV DNA for nasopharyngeal carcinoma. Oncotarget 2017;8:5292-308. [Crossref] [PubMed]
- Hong RL, Lin CY, Ting LL, et al. Comparison of clinical and molecular surveillance in patients with advanced nasopharyngeal carcinoma after primary therapy: the potential role of quantitative analysis of circulating Epstein-Barr virus DNA. Cancer 2004;100:1429-37. [Crossref] [PubMed]
- Leung SF, Lo YMD, Chan ATC, et al. Disparity of sensitivities in detection of radiation-naïve and postirradiation recurrent nasopharyngeal carcinoma of the undifferentiated type by quantitative analysis of circulating Epstein-Barr virus DNA1,2. Clin Cancer Res 2003;9:3431-4. [PubMed]
- Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2017;377:513-22. [Crossref] [PubMed]
- Hong Kong Cancer Registry. Nasopharyngeal cancer in 2018. Available online: https://www3.ha.org.hk/cancereg/
- Wong KCW, Hui EP, Lo KW, et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol 2021; Epub ahead of print. [Crossref] [PubMed]
- Blanchard P, Lee A, Marguet S, et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncol 2015;16:645-55. [Crossref] [PubMed]
- Ribassin-Majed L, Marguet S, Lee AWM, et al. What is the best treatment of locally advanced nasopharyngeal carcinoma? An individual patient data network meta-analysis. J Clin Oncol 2017;35:498-505. [Crossref] [PubMed]
- Au KH, Ngan RKC, Ng AWY, et al. Treatment outcomes of nasopharyngeal carcinoma in modern era after intensity modulated radiotherapy (IMRT) in Hong Kong: a report of 3328 patients (HKNPCSG 1301 study). Oral Oncol 2018;77:16-21. [Crossref] [PubMed]
- Chan ATC, Hui EP, Ngan RKC, et al. Analysis of plasma Epstein-Barr virus DNA in nasopharyngeal cancer after chemoradiation to identify high-risk patients for adjuvant chemotherapy: a randomized controlled trial. J Clin Oncol 2018;JCO2018777847. [Crossref] [PubMed]
- Kim KY, Le QT, Yom SS, et al. Current state of PCR-based Epstein-Barr virus DNA testing for nasopharyngeal cancer. J Natl Cancer Inst 2017;109:1-7. [Crossref] [PubMed]
- Le QT, Zhang Q, Cao H, et al. An international collaboration to harmonize the quantitative plasma Epstein-Barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin Cancer Res 2013;19:2208-15. [Crossref] [PubMed]
- Le QT, Colevas AD, O'Sullivan B, et al. Current treatment landscape of nasopharyngeal carcinoma and potential trials evaluating the value of immunotherapy. J Natl Cancer Inst 2019;111:655-63. [Crossref] [PubMed]
- Trevisiol C, Gion M, Vaona A, et al. The appropriate use of circulating EBV-DNA in nasopharyngeal carcinoma: Comprehensive clinical practice guidelines evaluation. Oral Oncol 2021;114:105128. [Crossref] [PubMed]
- Kanakry J, Ambinder R. The biology and clinical utility of ebv monitoring in blood. Curr Top Microbiol Immunol 2015;391:475-99. [Crossref] [PubMed]
- Chan KCA, Chu SWI, Lo YMD. Ambient temperature and screening for nasopharyngeal cancer. N Engl J Med 2018;378:962-3. [Crossref] [PubMed]
- Lam WKJ, Jiang P, Chan KCA, et al. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2018;115:E5115-24. [Crossref] [PubMed]
- Lo YMD, Chan KCA, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010;2:61ra91. [Crossref] [PubMed]
- Lo YMD, Han DSC, Jiang P, et al. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021;372:eaaw3616. [Crossref] [PubMed]
- Chan KCA, Jiang P, Sun K, et al. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci U S A 2016;113:E8159-68. [Crossref] [PubMed]
- Jiang P, Sun K, Tong YK, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci U S A 2018;115:E10925-33. [Crossref] [PubMed]
- Jiang P, Sun K, Peng W, et al. Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discov 2020;10:664-73. [Crossref] [PubMed]
- Han DSC, Ni M, Chan RWY, et al. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet 2020;106:202-14. [Crossref] [PubMed]
- Lam WKJ, Jiang P, Chan KCA, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun 2019;10:3256. [Crossref] [PubMed]
- Lam WKJ, Ji L, Tse OYO, et al. Sequencing analysis of plasma Epstein-Barr virus DNA reveals nasopharyngeal carcinoma-associated single nucleotide variant profiles. Clin Chem 2020;66:598-605. [Crossref] [PubMed]
- Hui KF, Chan TF, Yang W, et al. High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int J Cancer 2019;144:3031-42. [Crossref] [PubMed]
- Xu M, Yao Y, Chen H, et al. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat Genet 2019;51:1131-6. [Crossref] [PubMed]
- Lam WKJ, Chan KCA, Lo YMD. Plasma Epstein-Barr virus DNA as an archetypal circulating tumour DNA marker. J Pathol 2019;247:641-9. [Crossref] [PubMed]
Cite this article as: Lam WKJ, Lo YMD. Plasma Epstein-Barr virus DNA analysis for personalised management of nasopharyngeal carcinoma—current opportunities and challenges. Ann Nasopharynx Cancer 2022;6:4.


