
miRNA biomarkers for NPC diagnosis and prognosis
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs, typically 21–23 nucleotides long, that function in post-transcriptional gene regulation typically through translation inhibition and/or mRNA degradation (1). Since their discovery in Caenorhabditis elegans, miRNAs have been extensively investigated as master regulators of gene expression in a variety of animal, plant, and human models. MiRNA expression is frequently altered during cancer development, associated with dysregulated expression of a plethora of different miRNAs, their biogenesis or processing proteins, such as DROSHA and DICER1 (1). These miRNA alterations are potential diagnostic and prognostic biomarkers when assessing patient disease progression or guiding clinical management. Since their development in the early 2000s, several groups in both academia and industry have published papers and produced miRNA panels as potential products, which are available for clinical use, although these are still in clinical testing and not yet FDA-approved (2). Similarly, miRNA-based therapeutics are also in development for the treatment of a variety of human diseases. These clinical evaluations demonstrate the importance and potential of further miRNA research for clinical use, especially in the diagnosis and treatment of cancer.
Head and neck cancers (HNCs) are the seventh most common malignancy worldwide (3). One type of HNC is nasopharyngeal carcinoma (NPC), which arises from the nasopharyngeal mucosal lining often at the pharyngeal recess (4). The geographic distribution of NPC is notably unique wherein more than 70% of cases arise in east/southeast Asia, likely attributable to both genetic and environmental factors. NPCs are categorized into three subtypes (keratinising squamous, non-keratinising and basaloid squamous) (5). Of note, the non-keratinising subtype contributes to more than 95% of NPCs developing in endemic regions and is almost always associated with the Epstein-Barr virus (EBV), the most prevalent etiologic agent of NPC (6). The development of EBV-associated NPC is characterized by critical genomic alterations and aberrant signalling activities that ultimately ushers a normal, healthy epithelium towards malignancy (5). NPC miRNA expression profiling and signalling pathway elucidation suggest a dysregulated state caused by both cellular and viral-encoded miRNAs. While certain individual miRNAs may be informative of changes with respect to cellular growth, proliferation, metastases, and apoptosis for use in diagnostic or prognostic assessments, other specific miRNAs might guide the course of treatment by reflecting NPC resistance to radiotherapy (7-9) or chemotherapy (6,9-11). Advances in miRNA profiling, bioinformatics, and statistical analyses have permitted the identification of miRNA signatures with the potential to accurately diagnose or stratify patients into clinically relevant groups. Taken together, the identification of novel miRNA biomarkers and signatures may be crucial to clarify the underlying mechanisms of NPC pathogenesis and improve clinical outcomes.
miRNA biogenesis
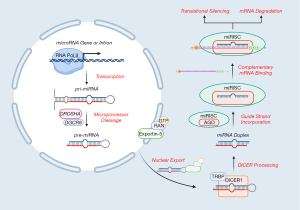
Human miRNA biogenesis begins in the nucleus with the transcription of primary miRNA (pri-miRNA) transcripts, predominantly by RNA polymerase II, although some RNA polymerase III-mediated transcription has also been reported (12,13) (Figure 1). Transcription most commonly occurs at designated miRNA promoters but has also been observed in the introns of protein-coding regions. These pri-miRNA transcripts can vary substantially in length and commonly adopt stem-loop secondary structures (14). The Microprocessor complex, comprised of the RNase III DROSHA and RNA-binding protein DiGeorge syndrome critical region 8 (DGCR8), subsequently cleaves the pri-miRNA at the base of the stem-loop structure to generate a ~60-70 nucleotide hairpin-shaped precursor miRNA (pre-miRNA) (6). Pre-miRNAs are then exported from the nucleus by exportin-5 and GTP-bound Ras-related Nuclear Protein (RAN-GTP) for further modification by the RNase III DICER1 and its associated cofactor transactivation-responsive RNA-binding protein (TRBP) (13). DICER1 processing produces a ~21-23 nucleotide miRNA duplex and one of the strands, termed the “guide strand”, is selectively integrated into the miRNA-inducible silencing complex (miRISC). Among other constituents, the major miRISC component is the Argonaute (AGO) protein (in humans, one of AGO 1–4) (12). The miRISC complex uses the incorporated guide strand to target mRNA transcripts and in turn, post-transcriptionally regulate its expression (13). A given miRNA guide strand may be complementary to numerous different transcripts and could possess several binding sites within a given mRNA (14). When partial complementarity exists between the guide strand and the target mRNA strand, AGO 1–4 can inhibit protein translation or direct the mRNA for non-specific degradation via cytoplasmic processing-bodies (P-Bodies) (14). Alternatively, when extensive or perfect complementarity exists between the guide strand and the target strand, the AGO2 protein may facilitate degradation of the mRNA through its “slicer” activity (Figure 1). In either case, these miRISC-mediated events primarily reduce the gene expression of the targeted mRNA transcript(s).
The functional roles of miRNAs in cancer
Given the broad repertoire of miRNAs identified in humans and their even more diverse array of target mRNAs, miRNAs are fittingly referred to as “master regulators” of gene expression. There are an estimated 2,000 miRNAs encoded in the human genome, which are further estimated to regulate at least one third of human genes (15,16). While global miRNA expression is typically downregulated in human cancer when compared to healthy tissues, aberrant expression of tumour-suppressor and oncogenic miRNAs can be critical in driving cancer initiation and subsequent progression (13). For example, two major oncogenic miRNAs are miR-21 and the miR-17-92 cluster, both of which are known to target tumour-suppressor genes involved in cell cycle regulation and apoptosis (17). Conversely, miRNAs of the let-7 family are regarded as tumour-suppressors, as they are known to target oncogenes, such as Myc and Ras family members, and are often downregulated in cancer cells (18). Similarly, downregulated expression of miRNA biosynthesis proteins (such as DROSHA and DICER1) has been associated with poorer clinical outcome in cancer patients (13). This may be in part due to an impaired DNA damage response, which decreases radiosensitivity (14). In EBV-associated NPC, both viral- and cellular-encoded miRNAs are pivotal in defining the cancer cell phenotype and predicting its development and response to treatment.
The role of EBV-encoded miRNAs in NPC
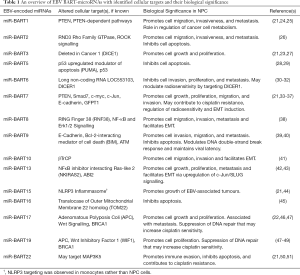
The EBV genome possesses two well-characterized regions, the BART and the BHRF1 genes, which encode 44 and 4 mature miRNAs (19), respectively. Notably, BHRF1 miRNA expression has not been observed in clinical NPC samples (20), while an overexpression of BART miRNAs has been well documented (21). BART miRNAs have also been detected in serum exosomes, vesicles, and soluble ribonucleoprotein complexes (22). Functionally, BART miRNAs are known to target an assortment of viral and cellular mRNAs to alter cellular proliferation, survival, and host immune response evasion (6,21,23). Consequently, BART-encoded miRNAs may be relevant as non-invasive biomarkers in the diagnosis and prognosis of NPC (Table 1).
Full table
A study investigating serum and plasma miRNA expression in NPC observed that miR-BART7-3p, 9-3p, and 13-3p were elevated in plasma, likely associated with extracellular vesicles or soluble ribonucleoprotein complexes (52,53). While the overexpression of these three miRNAs successfully distinguished NPC from healthy individuals and/or patients with non-NPC EBV-associated disease, circulating miR-BART13-3p was found to possess impressive specificity for NPC detection (97%) (52). MiR-BART13 has been previously implicated as a promoter of cell growth and metastases in NPC through an NFκB-dependent mechanism (42). Other investigations of plasma miR-BARTs similarly described an upregulation of miR-BART7 and miR-BART13, compared to healthy individuals (33,54). One study reported that plasma miR-BART7-3p, in conjunction with plasma EBV DNA, was associated with distant metastasis-free survival (DMFS), as patients with detectable abundances of both miR-BART7-3p and EBV DNA experienced poorer DMFS compared to patients with either factor independently or neither (54). In addition, miR-BART7 was identified as an in vitro promoter of proliferation (34), migration (33,35), and invasion (33), and could contribute to cisplatin resistance (36). Notably, miR-BART7-3p targets the tumour suppressor PTEN, and may augment PI3K/Akt signalling to facilitate nuclear accumulation of Snail and β-catenin to promote epithelial-to-mesenchymal transitions (EMT) (35,36) and metastases (35). MiR-BART7-3p targeting of Smad7 may promote transforming growth factor beta (TGF-β) signalling to mediate EMT and confer stemness, thereby enhancing the metastatic phenotype (36). One study reported an alternative mechanism whereby miR-BART-7 expression downregulates TGF-β1 through glutamine-fructose-6-phosphate transaminase 1 (GFPT1) targeting to increase radiosensitivity (37), although further investigation will be necessary to clarify these conflicting observations.
Elevated miR-BART2-5p levels have been detected in patient sera samples, and its abundance has been associated with poorer clinical outcomes, particularly as a result of elevated migration, invasion, and distant metastasis (26). The underlying mechanism may relate to miR-BART2-5p binding of RND3, a negative regulator of Rho signalling, which could indirectly promote ROCK signalling and increase the aggressiveness of cancer cells (26). MiR-BART2-5p upregulation has also been observed in other EBV-associated conditions (52). Nonetheless, miR-BART2-5p has demonstrated potential as a biomarker for early NPC diagnosis, and more studies are necessary to confirm its significance (55).
Overexpression of miR-BART10-3p is associated with downregulation of beta-transducin repeat containing E3 ubiquitin protein ligase (βTrCP), which may dysregulate downstream β-catenin and Snail, thereby promoting EMT in NPC (41). Indeed, miR-BART10-3p expression has been described as a negative prognostic marker, associated with lower disease-free survival (DFS) and overall survival (OS) rates (41). The negative correlation between miR-BART10-3p and βTrCP expression in NPC cells suggests that both molecules warrant further investigations as possible biomarkers.
Taken together, these findings demonstrate the biological significance of EBV-encoded BART miRNAs and their potential as informative biomarkers for NPC diagnosis and predictors for patient outcome. Among the plethora of BART miRNAs whose expression is altered in NPC, further investigation is necessary to clearly elucidate the functional underpinnings of these miRNAs and their role in NPC development (Table 1). In addition to identifying cellular factors and pathways that may be dysregulated as a consequence of BART miRNAs, developing miRNA signatures composed of several viral miRNAs could potentially unify the observations described in the literature, thereby enhancing the diagnostic efficacy of miRNA assays in NPC.
Although not the focus of this review, there are also EBV-encoded small RNAs (EBERs) transcribed in abundance from the EBV genome in NPC, which possess a variety of functional roles including regulation of cell growth and survival, host innate immunity and oncogenesis (56-58). EBERs, in addition to miRNAs and other interacting viral and host factors, underscore the complexity of molecular interactions underlying NPC.
Techniques employed in human genome-wide miRNA profiling and signature derivation
MiRNA profiling is typically conducted through a general technical workflow that spans from sample preparation and RNA extraction to subsequent profiling and analysis [reviewed in (59)]. Each step in this sequence necessitates care and quality control to ensure that the results generated by the assay are truly representative of the original sample and not obscured by ‘noise’. While both serum and plasma samples are commonly extracted from patients and directly profiled, tissue samples are often either freshly-extracted, frozen, or formalin-fixed paraffin-embedded (FFPE), thus introducing variability among samples that may affect profiling results (1,59). Numerous techniques exist for miRNA profiling, most prominently quantitative reverse transcription PCR (qRT-PCR), microarrays, and high-throughput RNA sequencing (RNA-seq), as well as other unique human miRNA assays such as Nanostring and Taqman Low-Density Arrays (TLDAs); however, each technique differs in its methodology (60-62). While qRT-PCR employs reverse transcription of miRNAs to cDNAs and subsequent qPCR to quantitatively monitor specific products generated, hybridization-based methods such as microarrays rely on fluorescent labelling and probe hybridization to quantify miRNAs (59). qRT-PCR, for example, is capable of measuring smaller miRNA panels with high sensitivity and specificity (62). Conversely, microarrays may measure greater numbers of miRNAs and more readily enable comparison of abundance between different samples (for example, a “healthy” vs. “diseased” sample), albeit with less sensitivity and specificity (62). A detailed discussion of miRNA profiling techniques is reviewed in Pritchard et al. (59) and Dave et al. (62).
Once profiling is completed, each dataset is subjected to statistical analyses and modelling, for example univariate/multivariate Cox analyses (63-65) or Risk Score calculations (63-65), dependent on the phenotypic variable being investigated to identify miRNAs that may be significant. Of note, certain bioinformatic investigations of miRNA signatures in NPC were conducted using datasets available from the Gene Expression Omnibus (GEO) database. The data produced and analyzed by Liu et al. (GSE32960) for example, was re-analyzed by Wang et al. and Zhang et al. (discussed below), and interestingly, yielded partially-overlapping miRNA signatures, which were subsequently validated using independent validation sets (63,64,66). Overall, while previous investigations of miRNAs in EBV-positive NPC sought to identify consistent biomarkers and miRNA signatures, they do not necessarily examine the same type of specimens or employ the same profiling techniques or statistical analyses throughout their workflow. The differences between their findings, as well as their methodologies, warrant consideration when selecting the most meaningful miRNA biomarkers for clinical and/or scientific use.
miRNA signatures in NPC
In NPC, miRNA expression profiles are known to be significantly different between healthy tissues and tumours, and furthermore, between tumour subtypes (6); hence, the diversity of tumours that may develop in NPC poses a significant challenge in accurately assessing patient outcomes (1). Rather than replacing conventional TNM staging, these miRNA signatures may be used in a complementary fashion to better assess patient prognosis and guide treatment selection (60,64,65). Table 2 summarizes all the published NPC miRNA signatures to date. Several of these studies employed pathway enrichment analyses to identify plausible targets of the miRNAs included in their signatures. These targets tended to be molecules or signalling pathways that might enhance cancer progression through alterations in cell cycle regulation, motility, survival, proliferation, or the extracellular matrix. While these signatures comprise a statistically significant selection of miRNAs with altered expression (either up- or downregulated) in NPC, it is also worth noting that individual miRNAs within these signatures have been implicated in the pathogenesis of NPC (6).

Full table
Liu et al. investigated miRNA expression profiles in 312 paraffin-embedded NPC samples (evenly randomized into a 156 sample training set and internal validation set), and 18 non-NPC samples using an 873 probe microarray and qRT-PCR, and subsequently, an independent set of 153 samples (64) (Table 2). A 5-miRNA signature was thus identified (miR-93, miR-142-3p, miR-29c, miR-26a, miR-30e) in which each miRNA was significantly associated with DFS, and as secondary outcomes, DMFS and OS (64). The prognostic value of this signature was greater when used in combination with TNM staging, while TNM staging performed less well alone, reiterating the enhanced prognostic value conferred by the addition of the miRNA signature (64). The dataset produced and analyzed by Liu et al. was uploaded to the GEO database (GSE32960) and was re-analyzed by Zhang et al. and Wang et al. to identify a partially-overlapping 4-miRNA signature and 3-miRNA signature, respectively (63,64,66). Zhang et al. analyzed the GSE32960 microarray dataset to identify 46 statistically significant differentially-expressed miRNAs (63). A weighted co-expression network was used in combination with univariate Cox regression analysis to identify 4 miRNAs (miR-142-3p, miR-150, miR-29b, miR-29c) that were most significantly associated with clinical traits such as DMFS, DFS, and OS, and also possessed the ability to classify patients into low-risk or high-risk groups (63). The 4-miRNA signature and its associated Risk Score generated by Zhang et al. was better able to predict the survival of NPC patients than TNM staging alone, similar to the signature identified by Liu et al. (63,64). While Zhang et al. identified potential mRNA targets of the miRNAs comprising their signature and consequently, protein pathways which may be altered, this study did not include a validation set to test the efficacy of their model with an independent set of samples (63).
Conversely, Wang et al. analyzed the GSE32960 dataset to produce a 3-miRNA signature (miR-142-3p, miR-29c, and miR-30e) which was subsequently validated using the independent GSE70970 dataset produced by Bruce et al. (60,66) (Table 2). This validation set demonstrated robustness of the signature in distinguishing survival between low-risk and high-risk groups (66). Of note, the 3-miRNA signature identified by Wang et al. overlapped with the 5-miRNA signature originally identified by Liu et al., while the 4-miRNA signature identified by Zhang et al. only partially overlapped. The independent validation that was conducted by Wang et al. and Liu et al. reiterates the significance of their overlapping miRNAs, particularly miR-142-3p, miR-29c, and miR-30e, and should therefore prompt further investigation into the significance of these miRNAs in NPC pathogenesis or potential for use as biomarkers.
Another study conducted by Liu et al. retroactively analyzed 512 serum specimens from newly-diagnosed, non-metastatic NPC patients pre-treatment (65) (Table 2). These samples were randomly allocated into a training set and validation set and four identified differentially-expressed miRNAs (miR-22, miR-572, miR-638, and miR-1234) were constructed into a statistically significant signature, which predicted poorer survival independent of clinical stage and elevated risk of distant metastasis (65). Notably, this 4-miRNA signature did not overlap with the previous 5-miRNA signature identified by Liu et al. and could thus, be an alternative means of assessing patient prognosis and guiding treatment selection, particularly among patients from the high-risk group who may benefit from more aggressive therapies (64,65).
Bruce et al. profiled the expression of 734 miRNAs from 135 diagnostic biopsy samples in a training set and 131 diagnostic biopsy samples in an independent validation set using the Nanostring Human miRNA Assay (60) (GSE70970 Dataset) (Table 2). The Cox Proportion Hazard Regression Model was used to develop a 4-miRNA signature that was associated with the risk of developing distant metastases, which was statistically significant in both the training (P<0.001) and validation (P=0.01) sets (60). Intriguingly, this signature could also discern ‘lower risk’ patients, who may be better treated with radiotherapy alone as opposed to chemoradiotherapy, thereby alleviating potentially unnecessary toxicity and the burden of treatment (60). When compared to the 5-miRNA signature identified by Liu et al., the signature identified by Bruce et al. seemed to perform better in terms of hazard ratio and statistical significance, with Bruce et al.’s model possessing a greater area-under-curve (AUC); however, due to extensive differences in the sample populations and methodologies employed between the studies, neither model can be described as superior over the other (60,64).
Zeng et al. analyzed serum miRNA expression using a TLDA and qRT-PCR to identify differentially-expressed miRNAs, and developed a 4-miRNA diagnostic signature (miR-17, miR-20a, miR-29c, miR-223) using the “Ct difference method” (61) (Table 2). When this diagnostic model was validated using an independent cohort of 74 NPC serum samples and 57 healthy serum samples, the sensitivity was 97.3% and specificity was 96.5% (61). The miRNAs identified in this study, particularly miR-17, miR-20a, and miR-29c, have been implicated in other signatures both derived from serum and other tissue sources (Table 2), suggesting that they may be useful as general biomarkers for NPC diagnosis across a variety of sample methods, although further investigations would definitely be necessary.
Zhang et al. identified a 7-miRNA plasma diagnostic signature by analyzing 200 NPC plasma samples and 189 healthy donors, which was found to be statistically significant following multi-phase validation (68) (Table 2). While six of these miRNAs were found to be significantly altered in tissue specimens, none of the seven miRNAs were significantly altered in plasma exosomes (68). A logistic regression statistical model was developed using the signature [Logit(P) equation] to distinguish NPC patients from healthy individuals, which possessed notable diagnostic ability based on receiver operating characteristic (ROC) curve analyses; however, the seven miRNAs comprising this signature were not significantly correlated with NPC prognosis (68). Nonetheless, further research into these seven identified miRNAs may shed light on molecular alterations in NPC tissues, and the differential miRNA expression/transport mechanisms that may distinguish miRNA profiles between tissues, exosomes, and plasma.
Wen et al. identified two miRNA signatures using microarray and qRT-PCR profiling of whole blood samples acquired from patients diagnosed with NPC, head-neck tumours (HNT), and healthy subjects (HSs) (67) (Table 2). A diagnostic model was constructed using an 8-miRNA signature in a training set of 84 NPC and 21 HSs samples, then validated using 36 NPC and 9 HSs independent samples (67). When applied to the validation set, this diagnostic model possessed notable accuracy, sensitivity, and specificity, when distinguishing NPC patients from HSs, and the AUC determined by ROC analyses was significant (P<0.01) (67). In an attempt to develop a signature that could distinguish NPC from other HNT, Wen et al. randomized a second training (84 NPC, 20 HNT, 22 HSs samples) and validation set (36 NPC, 10 HNT, 8 HSs samples), and identified 16 differentially-expressed miRNAs with diagnostic value for NPC, distinguishing from other HNT patients or HSs (67). When these miRNAs were constructed into a diagnostic model and validated, the 16-miRNA signature similarly possessed good sensitivity and specificity, with a statistically significant AUC (P<0.01) again, suggesting that this signature was able to adequately discern NPC cases from other HNT patients and HSs (67). Four miRNAs overlap between the 8-miRNA signature and the 16-miRNA signature (miR-4665-3p, miR-513b, miR-1908, miR-4284). Of these miRNAs, the variable importance plots for both signatures highlight miR-4665-3p as one of the most significant miRNAs in both models, while the non-overlapping miR-296-5p and miR-361-3p were most significant in the 16-miRNA signature (67). Further investigation into these miRNAs may provide important insights in distinguishing the molecular differences between healthy, HNT, and NPC tissues.
miRNA-associated cellular dysregulation in NPC
Numerous cellular miRNAs have been implicated for their role in altering NPC cellular phenotypes (69). Specific cellular targets of these miRNAs have been identified, in addition to roles in a variety of other malignancies. For example, miRNAs such as miR-34c (10), miR-200c (11), miR-205-5p (70), miR-223 (71), miR-296-5p (72), miR-379-5p (73) and miR-449b (74) may induce or suppress EMT. Others may be involved in the development or suppression of radioresistance, such as miR-17 (7), miR-20a-5p (8,75), miR-29c (9), miR-150 (76), and miR-205 (77), or chemoresistance, such as miR-29c (9,78), miR-34c (10), miR-200c (11) miR-449b (74), and miR-1278 (79). Notably, several miRNAs from the aforementioned signatures have been reported to adopt pathogenic roles, which have been subsequently validated in vitro. Given the plethora of dysregulated miRNAs in NPC, we will focus on the cellular role of miRNAs described in the various signatures.
Metastasis remains one of the major motivations in the development of miRNA signatures, given that many of the identified miRNAs regulate proliferation, migration, and invasion through a variety of signalling mechanisms. MiR-93 is one such example, having been reported to promote cell growth (80), proliferation (81,82), and invasion (80). Studies conducted in vitro have identified several targets of miR-93 including cyclin-dependent kinase inhibitor 1A (CDKN1A) (81), transforming growth factor beta receptor 2 (TGFβR2) (83), programmed cell death protein 4 (PDCD4) (82), and disabled homolog-2 (DAB2) (80). Collectively, miR-93 expression was upregulated in NPC cells and inversely correlated with the expression of these targets, which may favour proliferation as a consequence of aberrant cell cycle regulation and invasion through augmented TGF-β/Smad signalling, and in turn, PI3K/Akt signalling (83). These findings are consistent with the role of miR-93 in the signature proposed by Liu et al. (in 2012), wherein elevated expression of miR-93 contributed to higher risk scores, and in turn, increased the risk of distant-metastases (64).
As previously described, miR-29c, miR-30e, and miR-142-3p overlapped between several signatures, which were associated with distant metastasis, and treatment resistance. MiR-29c, in addition to its role as a regulator of chemosensitivity (9,78) and radiosensitivity (9), may suppress metastasis in a T-cell lymphoma invasion and metastasis-inducing protein 1 (TIAM1)-dependent manner (84). MiR-29c is significantly downregulated in NPC, which may thereby increase TIAM1 levels, and in turn, facilitate metastasis (84). This finding is consistent with the role of miR-29c in several signatures, as previously described, whereby lower levels of miR-29c were associated with poorer DMFS. Likewise, miR-30e-5p was also reported to inhibit migration and metastasis by targeting the metastasis-associated 1 (MTA1) protein (85), and ubiquitin-specific peptidase 22 (USP22) (86). Downregulated miR-30e-5p was associated with poorer prognosis, as would have been predicted by higher signature risk scores. While several signatures noted that elevated miR-142-3p would correspond to reduced risk scores, the literature is limited and controversial. One study proposes that downregulated or epigenetically silenced miR-142-3p, mediated by enhancer of zeste homolog 2 (EZH2)-recruited DNA methyltransferase 1 (DNMT1) in NPC cells may promote EMT and metastasis by upregulating zinc finger E-box-binding homeobox 2 (ZEB2) (87). Conversely, another study identified suppressor of cytokine signaling 6 (SOCS6) as a direct target of miR-142-3p, and postulated that miR-142-3p overexpression in NPC would promote tumorigenesis (88). This finding is conflicting given that several studies, including those that constructed signatures, noted miR-142-3p downregulation in NPC tissues was associated with poorer prognosis. Further research investigating expression profile alterations of individual miRNAs and their cellular consequences would be necessary to clarify this discrepancy. Regardless, the biological significance in metastasis of the miRNAs implicated in the proposed signatures is generally corroborated by independent studies noted in the literature.
TGF-β signalling has been implicated as a critical player in the development of various cancers, owing to its pleiotropic roles in cell proliferation, differentiation, migration, and survival (89). In EBV-positive cancers such as NPC, TGF-β signal transduction is further dysregulated by the interaction of both viral and host factors, including miRNAs (90) (see Section “The role of EBV-encoded miRNAs in NPC”). Notably, TGF-β functions in a biphasic manner during cancer development, acting as a tumour suppressor in the early stages of disease, but promoting tumor progression and aggressiveness in later stages, particularly through EMT (89,90). MiRNA-mediated regulation of TGF-β signalling and its downstream effectors may thus be a key contributor to NPC development and the successful prediction of clinical outcomes.
Our group conducted the global miRNA profiling in Bruce et al. (Table 2) and subsequently elucidated the biological significance of both miR-449b (74) and miR-34c (10) in NPC, two miRNAs further implicated in TGF-β signal transduction (Figure 2). Higher levels of miR-449b were reported in NPC cells, and patients presenting with elevated miR-449b expression experienced poorer 5-year OS (74). MiR-449b promoted chemoresistance in vitro, particularly cisplatin-resistance, by directly targeting transforming growth factor beta induced (TGFBI) protein, which resulted in downstream PTEN inactivation and Akt activation. Inhibition of either miR-449b or Akt was found to restore cisplatin sensitivity, as was observed when TGFBI was overexpressed.
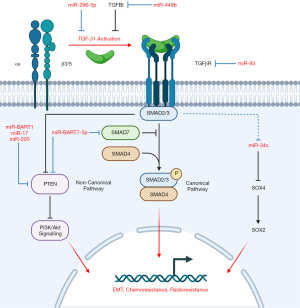
We also described a role of TGFBI-mediated inhibition of TGF-β1 signalling activity, in which TGFBI competes with extracellular latent TGF-β1 for binding of αvβ3/5 integrin (74). Downregulated TGFBI, as a consequence of miR-449b, would increase TGF-β1-integrin binding and subsequent TGF-β1 activation. Thus, canonical (Smad2/3) and non-canonical TGF-β signalling would be promoted and facilitate EMT and Akt activation, further enhancing chemoresistance. This outcome is consistent with the miRNA signature, as elevated miR-449b expression similarly corresponded to higher patient risk scores. These findings not only underscore the importance of both miR-449b and TGFBI as regulators of chemoresistance in NPC, but their potential value as biomarkers to inform treatment selection.
Our group identified that another miRNA from the Bruce et al. signature (60), miR-34c, targeted SOX4, a critical regulator in EMT, which was itself regulated by TGF-β1 further upstream (10). MiR-34c was found to be downregulated in NPC cells, relative to a normal nasopharyngeal cell line, which was simultaneously accompanied by elevated SOX4 and downstream SOX2 expression. EMT may be facilitated by SOX2/4 as suggested by increased expression of EMT markers in these cells (10). Furthermore, miR-34c inhibition conferred cisplatin-resistance to cells while its overexpression increased chemosensitivity. Thus, cells with low levels of miR-34c would likely possess a greater tendency to undergo EMT and develop chemoresistance, a finding consistent with the Bruce et al. signature (60). A study conducted by another group demonstrated that downregulated miR-34c-3p in NPC was associated with upregulated NOTCH1 and promoted cell growth, invasiveness, and EMT, further corroborating the relevance of miR-34c in NPC pathogenesis (91). While the precise mechanism of miR-34c downregulation could not be confirmed in our study, we proposed that TGF-β1 was a negative regulator of miR-34c that could alter SOX2/4 expression further downstream. Treatment with a TGF-β Receptor 1 inhibitor reduced SOX4 expression, and in turn, led to increased chemosensitivity.
When the two studies conducted by our group are considered in tandem (10,74), it is mechanistically plausible that miR-449b overexpression in NPC, and its corresponding downregulation of TGFBI and accumulation of TGF-β1, could downregulate miR-34c expression and upregulate SOX2/4 downstream to promote EMT and chemoresistance. Interestingly, the proposed miR-449b/miR-34c mechanism parallels the mechanism of miR-BART7-3p action described by Cai et al. (see Section “The role of EBV-encoded miRNAs in NPC”) wherein miR-BART7-3p similarly promotes canonical TGF-β signalling albeit by targeting regulatory Smad7 and promoting PI3K/Akt signalling through PTEN, ultimately conferring chemoresistance, stemness (as suggested by elevated stemness markers, including SOX2) and potentially EMT (36) (Figure 2). The convergence of these miRNAs, both cellular and viral-encoded, on TGF-β signalling, the PTEN/Akt axis and SOX2/4 expression, with their resulting consequences on chemoresistance and EMT, collectively underscores the biological significance of these pathways, and warrants further investigation into these molecules not only as biomarkers but potential therapeutic targets.
Challenges and future directions
Despite the great strides that have been achieved in understanding miRNA biology, there remains many challenges in elucidating the roles of miRNAs in NPC. The vast repertoire of miRNAs, both cellular and viral-encoded, is definitely daunting when attempting to identify the most significant players in the initiation or progression of this disease (6). Indeed, a plethora of individual miRNAs has been proposed as potential biomarkers in the literature. To this end, miRNA profiling techniques (59,62) and subsequent analyses aim to shed light on the most meaningful molecules or signatures that could be useful in the diagnosis, prognosis, or treatment selection of NPC patients. However, many studies remain hindered by differences in tissue type, tumour heterogeneity, and diverse profiling and analytical techniques employed (1). Consequently, the varying findings between studies may appear disparate and difficult to compare due to the differences that exist in methodologies. While some overlapping miRNAs exist between the signatures that have been highlighted in this review, the signatures themselves remain particularly diverse with their own unique miRNAs. Even so, the utility of these signatures is underscored by their significance with independent validations (60,61,64-68). Further investigation of the individual miRNAs comprising these signatures would be necessary to confirm their biological significance in NPC, especially in vivo studies to complement the multitude of miRNA investigations that have been conducted in vitro. At the time of this writing, no miRNA signatures have yet been identified with respect to EBV miR-BARTs, although future studies may provide additional insights given the high frequency of EBV-associated NPCs. Moving forward, unravelling the molecular events influenced by the intersection of miRNAs in NPC may help identify novel biomarkers, and additionally, therapeutic targets to improve clinical outcomes.
Conclusions
The biological significance of miRNAs in NPC is highlighted through their dual origin, both encoded by host cells or EBV, and the numerous cellular processes they influence as “master regulators” of the genome. Further investigations will be crucial to identify the most significant miRNA biomarkers and signatures in addition to their underlying relevance in NPC biology, potentially towards improved treatment, disease management, and potential therapeutic targets.
Acknowledgments
Funding: This work was supported by The Canadian Institutes of Health Research (#PJT - 153289), The Peter and Shelagh Godsoe Chair in Radiation Medicine, Canadian Cancer Society (#706321), The University of Toronto Faculty of Medicine, Princess Margaret Cancer Centre, The Princess Margaret Cancer Foundation, and the Ontario Ministry of Health.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maria Lung, Lawrence Young) for the series “NPC Biomarkers” published in Annals of Nasopharynx Cancer. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/anpc-21-4). The series “NPC Biomarkers” was commissioned by the editorial office without any funding or sponsorship. KWY reports that he is a co-applicant on grants for research on nasopharyngeal cancer, miRNAs, and related therapeutics. The granting agencies did not influence the writing of this manuscript. FFL reports that she is the principal applicant on grants for research on nasopharyngeal cancer, miRNAs, and related therapeutics, and she is also the Chair holder of the Peter and Shelagh Godsoe Chair in Radiation Medicine - a Hospital-University Named Chair established to support research. The granting agencies and the registered charity did not influence the writing of this manuscript. FFL was the co-chair of the 2018 Gordon Research Conference for Nasopharyngeal Carcinoma and served as an International Expert Advisor for a TRS-NPC project at The Chinese University of Hong Kong. FFL’s conference registration fee was waived and she received travel and accommodation support to attend the expert advisor meeting in Hong Kong. These agencies and their support did not influence the writing of this manuscript. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kwan JYY, Psarianos P, Bruce JP, et al. The complexity of microRNAs in human cancer. J Radiat Res (Tokyo) 2016;57:i106-11. [Crossref] [PubMed]
- Bonneau E, Neveu B, Kostantin E, et al. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019;30:114-27. [PubMed]
- Chow LQM. Head and Neck Cancer. N Engl J Med 2020;382:60-72. [Crossref] [PubMed]
- Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma. Lancet 2019;394:64-80. [Crossref] [PubMed]
- Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012-24. [Crossref] [PubMed]
- Spence T, Bruce J, Yip KW, et al. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol 2016;5:17. [Crossref] [PubMed]
- Hu Z, Zhou S, Luo H, et al. miRNA-17 promotes nasopharyngeal carcinoma radioresistance by targeting PTEN/AKT. Int J Clin Exp Pathol 2019;12:229-40. [PubMed]
- Huang D, Bian G, Pan Y, et al. MiR-20a-5p promotes radio-resistance by targeting Rab27B in nasopharyngeal cancer cells. Cancer Cell Int 2017;17:32. [Crossref] [PubMed]
- Zhang JX, Qian D, Wang FW, et al. MicroRNA-29c enhances the sensitivities of human nasopharyngeal carcinoma to cisplatin-based chemotherapy and radiotherapy. Cancer Lett 2013;329:91-8. [Crossref] [PubMed]
- Bissey PA, Teng M, Law JH, et al. MiR-34c downregulation leads to SOX4 overexpression and cisplatin resistance in nasopharyngeal carcinoma. BMC Cancer 2020;20:597. [Crossref] [PubMed]
- Yang J, Wu SP, Wang WJ, et al. A novel miR-200c/c-myc negative regulatory feedback loop is essential to the EMT process, CSC biology and drug sensitivity in nasopharyngeal cancer. Exp Cell Res 2020;391:111817 [Crossref] [PubMed]
- Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol 2017;1509:1-10. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Bruce JP, Liu F-F. MicroRNAs in nasopharyngeal carcinoma. Chin J Cancer 2014;33:539-44. [Crossref] [PubMed]
- Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3-14. [Crossref] [PubMed]
- de Rie D, Abugessaisa I, Alam T, et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol 2017;35:872-8. [Crossref] [PubMed]
- Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov 2016;6:235-46. [Crossref] [PubMed]
- Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-12. [Crossref] [PubMed]
- Chen SJ, Chen GH, Chen YH, et al. Characterization of Epstein-Barr Virus miRNAome in Nasopharyngeal Carcinoma by Deep Sequencing. PLoS One 2010;5:e12745 [Crossref] [PubMed]
- Cosmopoulos K, Pegtel M, Hawkins J, et al. Comprehensive Profiling of Epstein-Barr Virus MicroRNAs in Nasopharyngeal Carcinoma. J Virol 2009;83:2357-67. [Crossref] [PubMed]
- Wang Y, Guo Z, Shu Y, et al. BART miRNAs: an unimaginable force in the development of nasopharyngeal carcinoma. Eur J Cancer Prev 2017;26:144-50. [Crossref] [PubMed]
- Fan C, Tang Y, Wang J, et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer 2018;9:2852-64. [Crossref] [PubMed]
- Wang M, Yu F, Wu W, et al. Epstein-Barr virus-encoded microRNAs as regulators in host immune responses. Int J Biol Sci 2018;14:565-76. [Crossref] [PubMed]
- Cai L, Ye Y, Jiang Q, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun 2015;6:7353. [Crossref] [PubMed]
- Lyu X, Wang J, Guo X, et al. EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma. PLoS Pathog 2018;14:e1007484 [Crossref] [PubMed]
- Jiang C, Li L, Xiang Y-Q, et al. Epstein-Barr Virus miRNA BART2-5p Promotes Metastasis of Nasopharyngeal Carcinoma by Suppressing RND3. Cancer Res 2020;80:1957-69. [Crossref] [PubMed]
- Lei T, Yuen K-S, Xu R, et al. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer 2013;133:79-87. [Crossref] [PubMed]
- Choy EYW, Siu KL, Kok KH, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 2008;205:2551-60. [Crossref] [PubMed]
- Zheng X, Wang J, Wei L, et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J Virol 2018;92:e01022-18. [Crossref] [PubMed]
- He B, Li W, Wu Y, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis 2016;7:e2353 [Crossref] [PubMed]
- Wang D, Zeng Z, Zhang S, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell proliferation through the LOC553103-STMN1 axis. FASEB J 2020;34:8012-27. [Crossref] [PubMed]
- Tang J, Liu ZY, Tang Y, et al. Effects of Dicer1 targeted by EBV-miR-BART6-5p on biological properties and radiosensitivity of nasopharyngeal carcinoma. Hum Exp Toxicol 2021;40:977-93. [Crossref] [PubMed]
- Chan JYW, Gao W, Ho WK, et al. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res 2012;32:3201-10. [PubMed]
- Cai L, Li J, Zhang X, et al. Gold nano-particles (AuNPs) carrying anti-EBV-miR-BART7-3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget 2015;6:7838-50. [Crossref] [PubMed]
- Cai LM, Lyu XM, Luo WR, et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015;34:2156-66. [Crossref] [PubMed]
- Cai L, Long Y, Chong T, et al. EBV-miR-BART7-3p Imposes Stemness in Nasopharyngeal Carcinoma Cells by Suppressing SMAD7. Front Genet 2019;10:939. [Crossref] [PubMed]
- Gao W, Li ZH, Chen S, et al. Epstein-Barr virus encoded microRNA BART7 regulates radiation sensitivity of nasopharyngeal carcinoma. Oncotarget 2017;8:20297-308. [Crossref] [PubMed]
- Lin C, Zong J, Lin W, et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-κB and Erk1/2 pathways. J Exp Clin Cancer Res CR 2018;37:283. [Crossref] [PubMed]
- Hsu CY, Yi YH, Chang KP, et al. The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma. PLoS Pathog 2014;10:e1003974 [Crossref] [PubMed]
- Lung RW, Hau P, Yu KH, et al. EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma. J Pathol 2018;244:394-407. [Crossref] [PubMed]
- Yan Q, Zeng Z, Gong Z, et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget 2015;6:41766-82. [Crossref] [PubMed]
- Xu YJ, Zhou R, Zong JF, et al. Epstein-Barr virus-coded miR-BART13 promotes nasopharyngeal carcinoma cell growth and metastasis via targeting of the NKIRAS2/NF-κB pathway. Cancer Lett 2019;447:33-40. [Crossref] [PubMed]
- Huang J, Qin Y, Yang C, et al. Downregulation of ABI2 expression by EBV-miR-BART13-3p induces epithelial-mesenchymal transition of nasopharyngeal carcinoma cells through upregulation of c-JUN/SLUG signaling. Aging 2020;12:340-58. [Crossref] [PubMed]
- Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol 1950;2012:3795-9.
- Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival. PLoS Pathog 2015;11:e1004979 [Crossref] [PubMed]
- Hirai N, Wakisaka N, Kondo S, et al. Potential Interest in Circulating miR-BART17-5p As a Post-Treatment Biomarker for Prediction of Recurrence in Epstein-Barr Virus-Related Nasopharyngeal Carcinoma. PLoS One 2016;11:e0163609 [Crossref] [PubMed]
- Lung RW, Tong JH, Ip L, et al. EBV-encoded miRNAs can sensitize nasopharyngeal carcinoma to chemotherapeutic drugs by targeting BRCA1. J Cell Mol Med 2020;24:13523-35. [Crossref] [PubMed]
- Zhang Q, Luo D, Xie Z, et al. The Oncogenic Role of miR-BART19-3p in Epstein-Barr Virus-Associated Diseases. BioMed Res Int 2020;2020:5217039 [Crossref] [PubMed]
- Zhao Z, Liu W, Liu J, et al. The effect of EBV on WIF1, NLK, and APC gene methylation and expression in gastric carcinoma and nasopharyngeal cancer. J Med Virol 2017;89:1844-51. [Crossref] [PubMed]
- Lung RW, Tong JH, Sung YM, et al. Modulation of LMP2A Expression by a Newly Identified Epstein-Barr Virus-Encoded MicroRNA miR-BART22. Neoplasia 2009;11:1174-84. [Crossref] [PubMed]
- Liu Y, Jiang Q, Liu X, et al. Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3β/β-catenin signaling pathway. EBioMedicine 2019;48:386-404. [Crossref] [PubMed]
- Ramayanti O, Verkuijlen SAWM, Novianti P, et al. Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections. Int J Cancer 2019;144:2555-66. [Crossref] [PubMed]
- Gourzones C, Ferrand FR, Amiel C, et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients--evidence of non-exosomal transport. Virol J 2013;10:119. [Crossref] [PubMed]
- Lu T, Guo Q, Lin K, et al. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci 2020;111:1711-23. [Crossref] [PubMed]
- Jiang C, Chen J, Xie S, et al. Evaluation of circulating EBV microRNA BART2-5p in facilitating early detection and screening of nasopharyngeal carcinoma. Int J Cancer 2018;143:3209-17. [Crossref] [PubMed]
- Takada K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin Cancer Biol 2012;22:162-5. [Crossref] [PubMed]
- Takada K, Nanbo A. The role of EBERs in oncogenesis. Semin Cancer Biol 2001;11:461-7. [Crossref] [PubMed]
- Nanbo A, Takada K. The role of Epstein-Barr virus-encoded small RNAs (EBERs) in oncogenesis. Rev Med Virol 2002;12:321-6. [Crossref] [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [Crossref] [PubMed]
- Bruce JP, Hui ABY, Shi W, et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget 2015;6:4537-50. [Crossref] [PubMed]
- Zeng X, Xiang J, Wu M, et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 Combined as Non-Invasive Biomarkers in Nasopharyngeal Carcinoma. PLoS One 2012;7:e46367 [Crossref] [PubMed]
- Dave VP, Ngo TA, Pernestig A-K, et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest 2019;99:452-69. [Crossref] [PubMed]
- Zhang S, Yue W, Xie Y, et al. The four-microRNA signature identified by bioinformatics analysis predicts the prognosis of nasopharyngeal carcinoma patients. Oncol Rep 2019;42:1767-80. [Crossref] [PubMed]
- Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol 2012;13:633-41. [Crossref] [PubMed]
- Liu N, Cui RX, Sun Y, et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int J Cancer 2014;134:1359-68. [Crossref] [PubMed]
- Wang T, Wu J, Wu Y, et al. A novel microRNA-based signature predicts prognosis among nasopharyngeal cancer patients. Exp Biol Med (Maywood) 2021;246:72-83. [Crossref] [PubMed]
- Wen W, Mai SJ, Lin HX, et al. Identification of two microRNA signatures in whole blood as novel biomarkers for diagnosis of nasopharyngeal carcinoma. J Transl Med 2019;17:186. [Crossref] [PubMed]
- Zhang H, Zou X, Wu L, et al. Identification of a 7-microRNA signature in plasma as promising biomarker for nasopharyngeal carcinoma detection. Cancer Med 2020;9:1230-41. [Crossref] [PubMed]
- Chen HC, Chen GH, Chen YH, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer 2009;100:1002-11. [Crossref] [PubMed]
- Zhang P, Lu X, Shi Z, et al. miR-205-5p regulates epithelial-mesenchymal transition by targeting PTEN via PI3K/AKT signaling pathway in cisplatin-resistant nasopharyngeal carcinoma cells. Gene 2019;710:103-13. [Crossref] [PubMed]
- Gao L, Xiong X. MiR-223 inhibits the proliferation, invasion and EMT of nasopharyngeal carcinoma cells by targeting SSRP1. Int J Clin Exp Pathol 2018;11:4374-84. [PubMed]
- Chen M, Chen C, Luo H, et al. MicroRNA-296-5p inhibits cell metastasis and invasion in nasopharyngeal carcinoma by reversing transforming growth factor-β-induced epithelial-mesenchymal transition. Cell Mol Biol Lett 2020;25:49. [Crossref] [PubMed]
- Zhang F, Duan C, Yin S, et al. MicroRNA-379-5p/YBX1 Axis Regulates Cellular EMT to Suppress Migration and Invasion of Nasopharyngeal Carcinoma Cells. Cancer Manag Res 2020;12:4335-46. [Crossref] [PubMed]
- Bissey PA, Law JH, Bruce JP, et al. Dysregulation of the MiR-449b target TGFBI alters the TGFβ pathway to induce cisplatin resistance in nasopharyngeal carcinoma. Oncogenesis 2018;7:40. [Crossref] [PubMed]
- Zhao F, Pu Y, Qian L, et al. MiR-20a-5p promotes radio-resistance by targeting NPAS2 in nasopharyngeal cancer cells. Oncotarget 2017;8:105873-81. [Crossref] [PubMed]
- Huang Y, Tan D, Xiao J, et al. miR-150 contributes to the radioresistance in nasopharyngeal carcinoma cells by targeting glycogen synthase kinase-3β. J Cancer Res Ther 2018;14:111-8. [Crossref] [PubMed]
- Qu C, Liang Z, Huang J, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 2012;11:785-96. [Crossref] [PubMed]
- Huang L, Hu C, Chao H, et al. miR-29c regulates resistance to paclitaxel in nasopharyngeal cancer by targeting ITGB1. Exp Cell Res 2019;378:1-10. [Crossref] [PubMed]
- Zhao Y, Wang P, Wu Q. miR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol Cell Probes 2020;53:101597 [Crossref] [PubMed]
- Xu YF, Mao YP, Li YQ, et al. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett 2015;363:146-55. [Crossref] [PubMed]
- Zhang Y, Xu Z. miR-93 enhances cell proliferation by directly targeting CDKN1A in nasopharyngeal carcinoma. Oncol Lett 2018;15:1723-7. [PubMed]
- Sun J, Yong J, Zhang H. microRNA-93, upregulated in serum of nasopharyngeal carcinoma patients, promotes tumor cell proliferation by targeting PDCD4. Exp Ther Med 2020;19:2579-87. [Crossref] [PubMed]
- Lyu X, Fang W, Cai L, et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer 2014;13:51. [Crossref] [PubMed]
- Liu N, Tang LL, Sun Y, et al. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett 2013;329:181-8. [Crossref] [PubMed]
- Hu W, Yao W, Li H, et al. MiR-30e-5p inhibits the migration and invasion of nasopharyngeal carcinoma via regulating the expression of MTA1. Biosci Rep 2020;40:BSR20194309 [Crossref] [PubMed]
- Ma YX, Zhang H, Li XH, et al. MiR-30e-5p inhibits proliferation and metastasis of nasopharyngeal carcinoma cells by target-ing USP22. Eur Rev Med Pharmacol Sci 2018;22:6342-9. [PubMed]
- Li Y, He Q, Wen X, et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ 2019;26:1089-106. [Crossref] [PubMed]
- Qi X, Li J, Zhou C, et al. MiR-142-3p Suppresses SOCS6 Expression and Promotes Cell Proliferation in Nasopharyngeal Carcinoma. Cell Physiol Biochem 2015;36:1743-52. [Crossref] [PubMed]
- Syed V. TGF-β Signaling in Cancer. J Cell Biochem 2016;117:1279-87. [Crossref] [PubMed]
- Velapasamy S, Dawson CW, Young LS, et al. The Dynamic Roles of TGF-β Signalling in EBV-Associated Cancers. Cancers 2018;10:247. [Crossref] [PubMed]
- Xu X, Yan H, Zhang L, et al. Up-regulation of miR-34c-5p inhibits nasopharyngeal carcinoma cells by mediating NOTCH1. Biosci Rep 2020;40:BSR20200302 [Crossref] [PubMed]
Cite this article as: Fijardo M, Bissey PA, Yip KW, Liu FF. miRNA biomarkers for NPC diagnosis and prognosis. Ann Nasopharynx Cancer 2021;5:3.


